猪巨细胞病毒糖蛋白B基因克隆与序列分析
来源:wenku163.com 资料编号:WK1637907 资料等级:★★★★★ %E8%B5%84%E6%96%99%E7%BC%96%E5%8F%B7%EF%BC%9AWK1637907
资料介绍
猪巨细胞病毒糖蛋白B基因克隆与序列分析(7000字)
摘 要:猪巨细胞病毒(Porcine cytomegalovirus, PCMV)是一种影响猪健康、感染率极高病毒,也是一种以猪为供体的器官移植中不受欢迎的病毒。但迄今,对该病毒分子生物学特征的了解仍然十分有限。本论 文克隆并分析了来自临床病料的PCMV糖蛋白B(gB)基因,通过系统进化分析,首先发现PCMV已经进化为两个不同的基因型:B6株相关的基因型I和 OF-1株相关的基因型II,相应地,我们发现在基因型I PCMV毒株的gB蛋白中均存在的氨基酸序列“TA-P-S-D”,而在基因型II PCMV毒株的gB蛋白的相应位置这些氨基酸变为“ST-S-P-N”。这些结果表明PCMV已经进化为具有明显遗传差异两个基因型。而且,我们还发现这 两种基因型均在我国流行。
关键词:猪巨细胞病毒;gB基因;克隆;序列分析;
Cloning And Sequence Analysis Of Porcine Cytomegalovirus Glycoprotein B Gene
Abstract:Porcine cytomegalovirus (PCMV) is a ubiquitous and undesired pathogen for pig health and for use as organ donors in xenotransplantation. However, the molecular knowledge of the virus is still very lacking. In the present study, we sequenced and analyzed the complete nucleotide sequences of glycoprotein B (gB), gene of PCMV strains from clinic samples in China. For the first time, based on the phylogenetic analysis of complete nucleotide sequences of gB, PCMV strains were divided into two distinct genogroups: B6-related clade I and OF1-related clade II, and, correspondingly, special amino acid alterations of “TA-P-S-D” to “ST-S-P-N” were found in gB protein of B6-related clade I and OF1-related clade II strains, respectively, indicating the existence of subgroups of PCMV with obvious genetic differentiation. Moreover, the PCMV strains in the two clades are discovered to be co-circulating in China.
Key words:Porcine cytomegalovirus; gB genes; Cloning; Sequence analysis
目 录
摘要…………………………………………………………………………………1
关键词………………………………………………………………………………1
1前言………………………………………………………………………………2
2材料与方法………………………………………………………………………4
2.1样本…………………………………………………………………………4
2.2 DNA制备…………………………………………………………………5
2.3主要试剂……………………………………………………………………5
2.4引物的设计与合成………………………………………………………5
2.5 gB基因片段PCR扩增……………………………………………………5
2.6序列分析……………………………………………………………………6
3结果与分析…………………………………………………………………………6
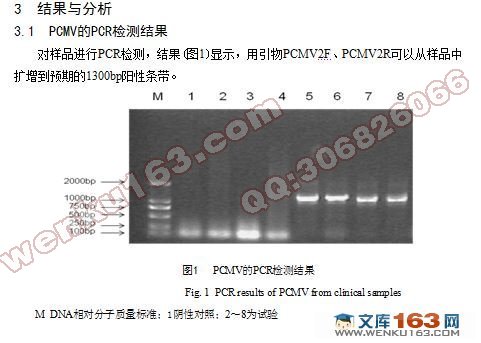
3.1 PCMV的PCR检测结果……………………………………………………6
3.2 gB基因序列分析……………………………………………………………6
4讨论…………………………………………………………………………………8
5结论…………………………………………………………………………………9
参考文献………………………………………………………………………………9
致谢…………………………………………………………………………………11
|